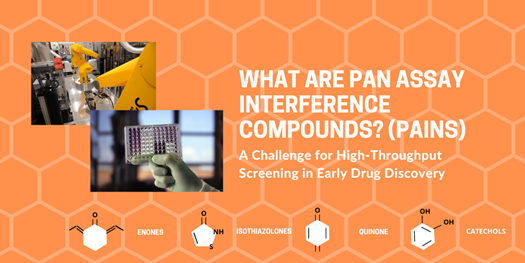
Legend (6, 7). Pan-assay interference compounds that produce a false-positive readout in a high-throughput screen. For an apparent hit to be considered a true hit, the compound must bind directly to the binding site of the target protein and produce the desired effect. PAINS, however, are tagged as hits due to interference with the biological or biochemical properties of the assay. It is critical for researchers to identify PAINS in their screens in order to eliminate them from further consideration. Otherwise, an exorbitant amount of time and money cant be spent on a compound that cannot be developed into a drug. Picture 1 is a robot used for high-throughput screens; Picture 2 is an assay plate loaded with samples; Pictures 3-6 (designed by the author) are the chemical structures of some of the most common PAINS. Image created using Canva Pro.
Summary
Drug Discovery Process
The preclinical process of target-based drug discovery is composed of three phases. The first is the discovery stage in which researchers identify a valid therapeutic target, develop a target-based pharmaceutical assay, and screen a large number of potential therapeutic agents. The second is the lead optimization stage in which researchers engineer chemical analogs of the lead molecules and perform assays to test their activity. In order for the optimized lead to become a drug candidate for clinical study, researchers must determine whether the optimized lead has sufficient activity for the target of interest and does not exhibit non-druglike properties. The final stage is the clinical development phase in which researchers must select the most appropriate lead for testing in clinical trials. While the drug discovery process appears to be a simplified series of steps, it requires years of endurance and commitment on behalf of the researchers in hopes of identifying a promising drug candidate (1).
HTS Triaging
One of the most challenging parts of drug discovery is the identification of small molecules that directly bind to the target of interest and affect its biological activity (2). The advent of high-throughput screening (HTS) has enabled medicinal chemists to screen for hundreds of thousands of compounds in a time and resource efficient manner and identify hits with a desired effect on the target of interest (2). An important aspect of HTS is HTS triaging in which hits are tested for their experimental validity and are eliminated when undesirable activity is detected (3). Compounds with undesirable activity can include either molecules that are active but impact multiple targets through the mechanism of action (MoA) of interest or molecules that bind the target through an undesirable MoA (3). However, there also exist compounds that yield false assay readouts about the effect of the compound of the target molecule (3). These chemical molecules are called pan-assay interference compounds, or PAINS (1)
PAINS
PAINS are chemical compounds that often yield false positives in HTS. In fact, it is estimated that a typical academic screening library will contain roughly 5-12% of PAINS (2). There are approximately 400 structural classes of PAINS, with more than half of them falling under 16 easily identifiable groups (2). Some of the most common classes of PAINS include toxoflavin, isothiazolones, enones, curcumin, hydroxyphenyl hydrazones, ene rhodanine, phenol sulphonamides, and quinones and catechols (2). PAINS function as reactive chemicals rather than target-specific drugs and are tagged as hits primarily because their substructures have the ability to interfere in biochemical assays (4). In fact, some mechanisms of interference include reactivity with biological and bioassay reagents, photoreactivity with proteins, metal chelation that affects proteins or assay reagents, redox activity, and photochromic properties that may impact absorption and fluorescence (4).
One noteworthy example of the prevalence of PAINS comes from Dr. Michael Walters at the University of Minnesota (5). In 2015, the Walters lab was interested in a histone acetyltransferase Rtt109 as a potential antifungal target (5). The team developed a HTS that detects thiol-containing fluorescent byproduct from an enzymatic reaction. The HTS yielded 1500 apparent hits from a screen of over 225,000 compounds (5). However, further studies revealed that only 3 of the 1500 apparent hits turned out to be true hits (5). Computational methods such as electronic filters have been developed and are widely used during the HTS triage process to quickly identify PAINS substructures and exclude them from further analysis (3). Skeptics, however, condemn electronic filters and criticize them as overly simplistic processes that may eliminate a potential compound from consideration and may classify a useless compound as promising (4). Nonetheless, research groups continue their efforts to develop an improved set of PAINS filters in order to optimize the drug discovery process.
Concluding Remarks
It is critical to identify PAINS during the discovery process of drug development. If PAINS are not identified early on in drug discovery, medicinal chemists will spend a significant amount of time and money trying to optimize non-progressable compounds and their efforts to develop an effective therapeutic for a disease will be futile (5). Moreover, repeated identification of the same molecules as promising hits in scientific publications will skew the literature by revalidating PAINS as good drug leads (2).
References
- Kenakin TP. 2014. The Drug Discovery Process. In: A Pharmacology Primer. Elsevier. p. 1795–1802.
- Baell J, Walters MA. 2014. Chemistry: Chemical con artists foil drug discovery.Nature 513:481–483.
- David L, Walsh J, Sturm N, Feierberg I, Nissink JWM, Chen H, Bajorath J, Engkvist O. 2019. Identification of Compounds That Interfere with High-Throughput Screening Assay Technologies.ChemMedChem 14:1795–1802.
- Erlanson DA. 2015. Learning from PAINful lessons.Journal of Medicinal Chemistry 58:2088–2090.
- Baell JB, Nissink JWM. 2018. Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017—Utility and Limitations.ACS Chemical Biology 13:36–44.
- National Institute of Health (NIH) . 2009 Aug 28. NCATS Robot Lab. Creative Commons; [accessed 2021 May 3]. https://search.creativecommons.org/photos/028bd7bb-71bd-47d9-9497-e2501035b31e
- National Institute of Arthritis and Musculoskeletal. 2009 Aug 28. Protein Assay Plate to be Analyzed by the Spectrometer. Creative Commons; [accessed 2016 Jan 4]. https://search.creativecommons.org/photos/a3079eac-4cdc-4f0c-9b93-d4f70bc17e02