An Analysis of High Throughput Screening with a Focus on Phenotypic Screens
Author: Anonymous
Learning Objectives
LO1: Describe the basics of phenotypic high throughput screening.
LO2: Summarize the differences between phenotypic and target-based high throughput screening.
LO3: Identify what has made phenotypic screens more prevalent in recent years.
LO4: Describe the specifics of a popular phenotypic screening method.
Graphical Abstract
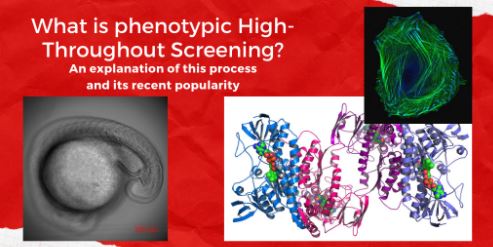
Legend. High Throughput (HTP) screening applies ideas such as miniaturization and automation to common biotechnological processes to allow researchers to screen upwards of hundreds of thousands of compounds at once. This greatly increases the speed at which new potential drugs can be found. HTP screens are typically done with one of two general methods. The first is a phenotypic screen, and the second is a target-based screen. The top right image shows a cell, representing the focus of phenotypic screens on the effect of a compound on the morphology of an entire cell. The bottom right image represents how target-based screens focus on protein affinity of compounds. Finally, the bottom left image shows a zebrafish embryo. Phenotypic screening can also be done on entire organisms, and zebrafish embryos like the one shown are one of the most common test organisms. All images from http://creativecommons.org and are used for educational purposes only. Image created using Canva Pro.
Background
High throughput (HTP) screening is a series of processes developed to increase the speed at which new drugs, or potential drugs can be developed. Researches engaged in HTP screening use automation and miniaturization of common biotechnology processes to be able to screen more than 10,000 compounds at once, in an effort to find which of those compounds are biologically active [1]. In searching for those biologically active compounds, or “hits”, there are two general types of HTP screening: phenotypic screens and target-based screens. These methods have different benefits and drawbacks, and as such are often preferred in different situations. Target-based screens are based around the premise of testing and validating already hypothesized molecular interactions. To accomplish this, researchers first identify a gene that codes for the protein of interest that is involved in a process, and then can screen for compounds with affinity for that protein [2]. This contrasts with phenotypic screens, where the effect of a compound is considered first, and the screening process searches the library of compounds for one that alters the phenotype of a cell or organism [3]. A key benefit of phenotypic screens is the lack of bias present when discovering whether a compound is active, due to the lack of need to validate the identity of the compounds’ target. However, phenotypic screens usually require much more downstream analysis of results to denoise signals, which can, due to the additional time and technology needed to accomplish this, greatly reduce the throughput of this type of screen. Target based screens require less verification of the results downstream, and do not need researchers to acquire relatively large amounts of cells like a phenotypic screen would [4].
The advantages of target-based screening, in combination with advancements in genomics in the late 1980’s made target-based screens more popular than phenotypic screens during that time. However, this trend has been reversing in recent years, as phenotypic screens have begun to become more popular in the search for new compounds or drugs [5]. One major reason for this resurgence, is that recently target-based screening has started to be seen as too restrictive. Specifically, it is becoming a more common belief in the drug development field that the pharmacology of compounds cannot accurately be described by looking at just one target, as in a target-based screen. The thought processes is thus, that a target-based screen vastly over simplifies the biological pathways involved in a disease, for example, and that by only looking at one specific target, a researcher will not properly understand the biology of the illness. In light of this, scientists have begun to look back and have found that the switch to a more target-based centric view of screening may have in fact, actually decreased the rate at which novel hits, and therefore drugs, had been found in certain situations in the decades before this recent resurgence of phenotypic screens [6]. One specific example is the pharmaceutical company GlaxoSmithKline, which in recent years has begun to move back towards the use of phenotypic screening due to advancements to chemical proteomics, and has since seen an increase in the speed at which they can finish their projects [3].
Finally, one especially common phenotypic screening method is flow cytometry. The method fires a laser and light scatter of cells is measured as they pass through it. From this, a researcher can very quickly analyze vast, complicated groups of cells based on specific testing criteria they set beforehand [7,8]. This method is widely used for both microbiology, and analysis of much larger animal cells, and shows off very well the benefits of HTP techniques, as it allows researchers to look at the expression of up to 10,000 cells a second [8].
References
[1] Attene-Ramos, Matias S, et al. “High Throughput Screening.” Encyclopedia of Toxicology, 3rd ed., Elsevier, 2014, pp. 916–917.
[2] Lage, Olga Maria et al. “Current Screening Methodologies in Drug Discovery for Selected Human Diseases.” Marine drugs vol. 16,8 279. 14 Aug. 2018, doi:10.3390/md16080279
[3] Kotz, J. Phenotypic screening, take two. Science-Business eXchange 5, 380 (2012). https://doi.org/10.1038/scibx.2012.380
[4] Technology Networks, Joanna Owens. “Phenotypic versus Target-Based Screening for Drug Discovery.” Drug Discovery from Technology Networks, Technology Networks, 9 Apr. 2020, www.technologynetworks.com/drug-discovery/articles/phenotypic-versus-target-based-screening-for-drug-discovery-300037.
[5] Mason, Laura Elizabeth. “The Role of Phenotypic Screening in Drug Discovery.” Drug Discovery from Technology Networks, 25 Oct. 2017, www.technologynetworks.com/drug-discovery/articles/the-role-of-phenotypic-screening-in-drug-discovery-293422.
[6] Swinney, D., Anthony, J. How were new medicines discovered?. Nat Rev Drug Discov 10, 507–519 (2011). https://doi.org/10.1038/nrd3480
[7] “Introduction to Flow Cytometry.” Abcam.com, Abcam, 15 Sept. 2020, www.abcam.com/protocols/introduction-to-flow-cytometry.
[8] Brehm-Stecher, Byron F. “Flow Cytometry.” Encyclopedia of Food Microbiology, 2nd ed., Elsevier, 2014, pp. 943–953.
Audio Recordings
Questions
- What does High Throughput screening entail, and how does it aid research?
- HTP screening takes standard biotechnological and drug development techniques and automates and miniaturizes them. This allows more compounds to be screened for potential hits in a shorter amount of time, allowing a sped-up drug discovery process.
- What are some key differences between Phenotypic and Target-based screens?
- One key difference is simply the focus of the two types of screens. Phenotypic screens focus on the overall morphology of a cell or an organism, and how it changes when exposed to compounds, while a target-based screen looks first at the protein affinities of different compounds. Additionally, while target-based screens try to answer existing hypotheses, phenotypic screens are not so restricted, and can be used to identify new biological interactions.
- What is the main factor that has caused the reliance on target-based screens to decrease recently, and caused phenotypic screens to rise in popularity among researchers?
- In general, the reason this switch has occurred is due to a switch of philosophies among drug-development and biotechnology researchers. Improvements in genomic technologies and methodologies in the 1980’s 1990’s, and early 2000’s made target screens more popular because it was much easier to identify a protein of interest, and therefore create a hypothesis about the cause of an illness, or condition. However, recently, meta-research has begun to show that these screens are not as much more effective as originally thought. Due to this, the primary philosophy among many of those researchers has begun to be that because of the more in depth understanding of all the biology involved with many complicated conditions that can be gained by phenotypic screens, that those are now the way to go.